https://doi.org/10.1007/s00392-025-02625-4
1Universitätsklinikum Tübingen Innere Medizin III, Kardiologie und Angiologie Tübingen, Deutschland; 2Universitätsklinikum Tübingen Innere Medizin III, Kardiologie und Kreislauferkrankungen Tübingen, Deutschland; 3Uniklinik RWTH Aachen Med. Klinik I - Kardiologie, Angiologie und Internistische Intensivmedizin Aachen, Deutschland
Background and aims
During cardiac remodeling disruption of the extracellular matrix (ECM) plays a pivotal role. The ECM not only provides a structural scaffold, but also facilitates molecular, electrical, and mechanical signaling. During early remodeling, a tachytrophic microenvironment forms, marked by high metabolic activity, increased signaling, and rapid matrix turnover. In contrast, once established, fibrosis and scarring form a bradytrophic microenvironment with impaired cell signaling and electrical connectivity, as well as an accompanying reduced metabolic activity.
EMMPRIN (CD147), a glycoprotein receptor, is critically involved in both inflammatory and fibrotic remodeling processes, as well as cellular energy metabolism.
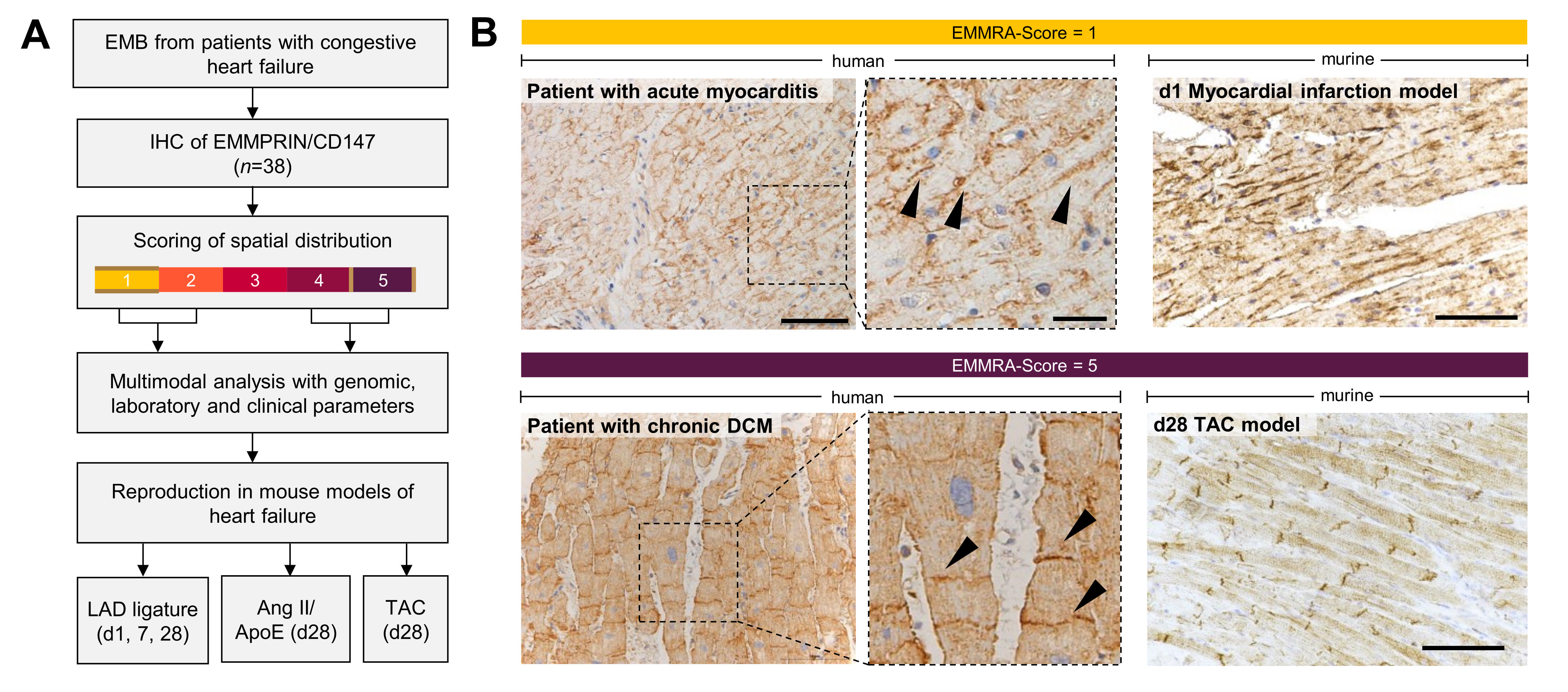
In this study, we describe a novel histological pattern of EMMPRIN localization, the EMMRA (EMMPRIN ReArrangement)-sign, found in human endomyocardial biopsies (EMBs) and murine heart failure models. We demonstrate that EMMPRIN exhibits differential polarity depending on the ongoing remodeling process: longitudinal expression along the cardiomyocyte membrane correlates with the tachytrophic inflammatory phase, while its localization at the shorter cell sides is associated with the bradytrophic fibrotic phase of “burnt-out” cardiomyopathies and established fibrosis.
Methods
EMBs from patients with diverse types of cardiomyopathies were examined, focusing on those with clear histological EMMPRIN localization patterns (n=38). Transcriptomic analyses using the NanoString® nCounter assay with 594 inflammation-related genes were conducted to explore the association of EMMPRIN localization with myocardial inflammation and fibrosis. Publicly available proteomic and genomic datasets were used to explore the quantitative regulation of EMMPRIN and its gene BSG in cardiomyopathies.
Human analyses were paralleled by murine heart failure models with acute (inflammatory) and chronic (fibrotic) remodeling.
Results
Distinct EMMPRIN spatial patterns were observed in human and murine hearts, while protein and gene expression levels of EMMPRIN were consistent across cardiomyopathies.
We observed large discrepancies between MRI diagnosis, molecular and (immune-) histological EMB pathology diagnosis and the actual clinical presentation of the patient. Surprisingly, most patients presenting with acute or subacute heart failure symptom onset displayed an EMMPRIN localization at the long cardiomyocyte side. In contrast, patients with known chronic heart failure or gradual symptom onset showed a modal EMMRA-Score of 5, which was significantly higher compared to acute (P=0.003794) or subacute (P=0.007504) onsets.
Transcriptomic analysis of EMBs suggested strong and highly significant upregulation of inflammation-associated genes in patients with EMMRA-Scores 1-2 compared to those with EMMRA-Scores 4-5.
We aimed to reproduce the findings in murine heart failure models where the disease onset is clearly defined. Here, EMMRA-Score 1 predominated post-LAD ligation in early phases, while chronic models with little or no inflammation (TAC model, Ang II/ApoE-/- model) showed mostly EMMRA-Scores of 5.
Conclusion
EMMPRIN rearrangement reflects tachytrophic (inflammatory) versus bradytrophic (fibrotic) cardiac states. The EMMRA-sign may therefore serve as a histological marker for cardiomyopathy progression tendencies, identifying stages potentially responsive to interventions before irreversible fibrosis sets in.